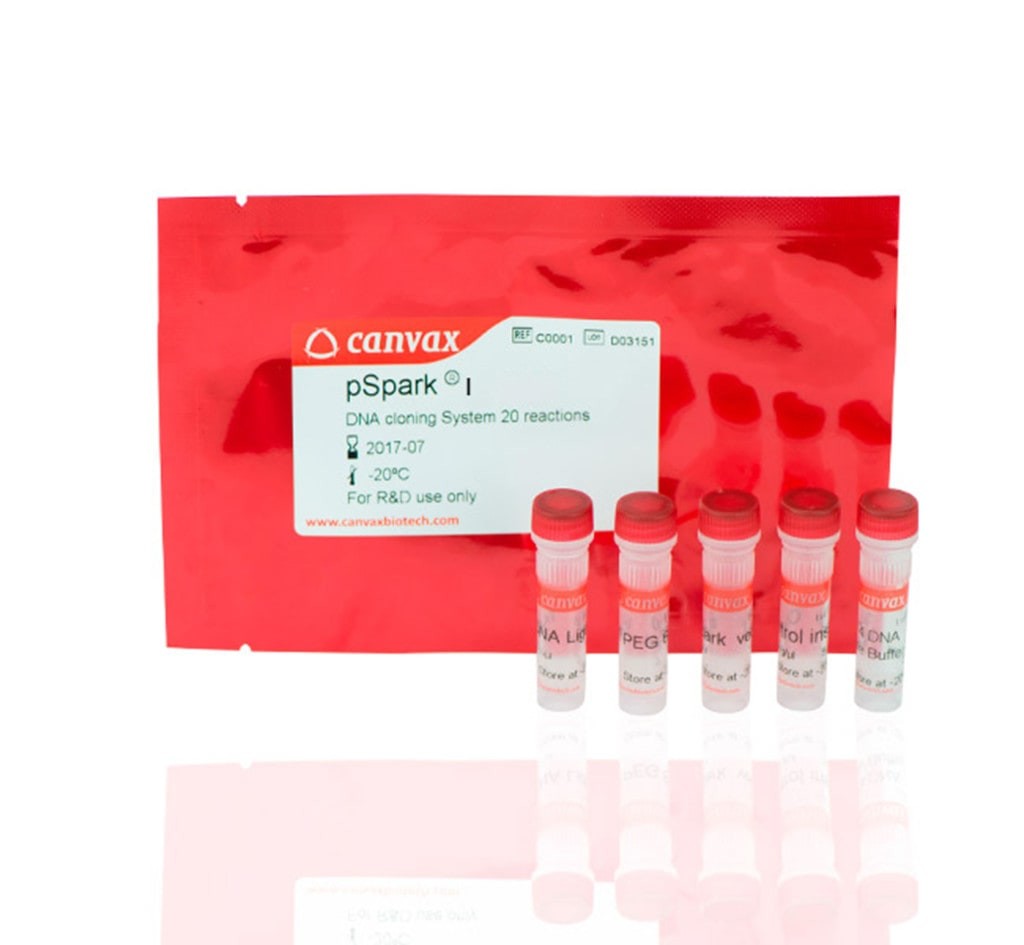
pSpark® IV
For highly Efficient, Stable & Powerful Cloning under transcription-free conditions
pSpark® IV is a highly efficient, accurate and easy-to-use DNA cloning system that exploit its very low background feature for the expression of toxic genes under transcription-free conditions. In this vector, the lac promoter has been eliminated and therefore blue/white screening is not allowed (alpha-peptide coding region remains and you can find blue colony). The vector is ideal for cloning genes that produce toxic polypeptides by transcription/ translation.
Price: 184.68 €
Detailed information:
Advantages & Features
- Unprecedented high cloning efficiency: more than 2,500 positive colonies expected under optimal conditions.
- Transcription-free.
- Easy-to-use: eliminate screening of recombinants due to its minumum background (lower than 1%).
- High stability: eliminates cloning bias or pitfalls.
- Time-saving protocol: avoids any step required after PCR, just 19 minutes from PCR to plating.
- Powerful: obtain 5-fold more positive colonies using 10-fold less DNA insert.
- Great versatility: compatible with any protocol, proofreading Polymerase, Competent Cells, ligation time or primers.
- Sensitive: clone from 50 bp insert to up to 14 kb with just 5 ng per kb of insert.
- Cost avoidance: removes expensive primer phosphorylation use.
- Eliminates positive selection vector.
Specifications
Includes
– 20 rxn pSpark® IV (20 ng/µL)
– 20 µL T4 DNA Ligase (5 Weiss U/µL)
– 100 µL T4 DNA Ligase Buffer (10x)
– 150 µL PEG 6000 (10x)
– 5 µL Insert Control 1 kb (20 ng/µL)
Applications
- Cloning of high fidelity PCR amplified products.
- Production of ssDNA.
- In vitro transcription from T7/SP6 dual-opposed promoters.
- Cloning of toxic genes.
Tables & Figures
t
Quality Control
- Functional test using a 1.0 kb PCR fragment.
Advice
- Recommendations: All pSpark® DNA cloning vectors are stable for at least 1 month at 4 °C and even at 20 – 25 °C for up to 2 days although storage temperatures above -20 °C are not recommended. In case of incident, such as a power failure, stored vectors should be tested with the supplied control insert before considering discarding it. However, please note that T4 DNA Ligase is extremely temperature-sensitive and storage temperatures above -20 °C inactivates the enzyme.
Storage, Shipping & Guarantee
- Shipped in: Gel Pack.
- Storage: -20º C (NON Frost-Free Freezer).
Citations
- Kristína Papayova, Lucia Bocanova, Vladena Bauerova, Jacob Bauer, Nora Halgasova, Maria Kajsikova, Gabriela Bukovska, (2024) From sequence to function: Exploring biophysical properties of bacteriophage BFK20 lytic transglycosylase domain from the minor tail protein gp15, Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics.
- Paullada‐Salmerón, J. A., Wang, B., & Muñoz‐Cueto, J. A. (2023). Spexin in the European sea bass, Dicentrarchus labrax: Characterization, brain distribution, and interaction with Gnrh and Gnih neurons. Journal of Comparative Neurology, 531(2), 314-335.
- Huertas-García, A. B., Guzmán, C., Tabbita, F., & Alvarez, J. B. (2022). Allelic variation of puroindolines genes in Iranian common wheat landraces. Agriculture, 12(8), 1196.
- Gómez Gil, L. (2022). Structure and dynamics of chromosomes: role in the genomic and pathogenic plasticity of Fusarium oxysporum.
- Oliveras, À., Camó, C., Caravaca-Fuentes, P., Moll, L., Riesco-Llach, G., Gil-Caballero, S., … & Planas, M. (2022). Peptide Conjugates Derived from flg15, Pep13, and PIP1 That Are Active against Plant-Pathogenic Bacteria and Trigger Plant Defense Responses. Applied and Environmental Microbiology, e00574-22.
- Morales de los Ríos Martín, L. (2022). Influencia del transportador de nitrato NRT1. 4/NPF6. 2 en la nutrición de potasio en Arabidopsis thaliana.
- Oliveras, À., Camó, C., Caravaca-Fuentes, P., Moll, L., Riesco-Llach, G., Gil-Caballero, S., … & Planas, M. (2022). Peptide Conjugates Derived from flg15, Pep13, and PIP1 That Are Active against Plant-Pathogenic Bacteria and Trigger Plant Defense Responses. Applied and Environmental Microbiology, e00574-22.
- Sueiro, R., Lamas, J., Palenzuela, O., Gulias, P., Diez-Vives, C., García-Ulloa, A., & Leiro, J. M. (2022). A real-time quantitative PCR assay using hydrolysis probes for monitoring scuticociliate parasites in seawater. Aquaculture, 738303.
- de los Ríos, Laura Morales, et al. “The Arabidopsis protein NPF6. 2/NRT1. 4 is a plasma membrane nitrate transporter and a target of protein kinase CIPK23.” Plant Physiology and Biochemistry (2021).
- Oliveras Rovira, Àngel, et al. “Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens.” PLoS ONE, 2018, vol. 13, núm. 7, p. e0201571 (2018).
- Foix, Laura, et al. “Prunus persica plant endogenous peptides PpPep1 and PpPep2 cause PTI-like transcriptome reprogramming in peach and enhance resistance to Xanthomonas arboricola pv. pruni.” BMC Genomics 22.1 (2021): 1-18.
- Jiménez Muñoz, R. (2020). Desarrollo de herramientas fisiológicas y genómicas para mejorar la calidad postcosecha del fruto de calabacín.
- Arévalo Díaz, S. (2020). Proteins influencing the septal junctions in heterocystous cyanobacteria.
- Brito, J., Valle, A., Almenglo, F., Ramírez, M., & Cantero, D. (2020). Characterization of eubacterial communities by Denaturing Gradient Gel Electrophoresis (DGGE) and Next Generation Sequencing (NGS) in a desulfurization biotrickling filter using progressive changes of nitrate and nitrite as final electron acceptors. New Biotechnology.
- de Felipe González, A. P. (2019). Resolución de la sinonimia Philasterides dicentrarchi/Miamiensis avidus. Caracterización genética, serológica y bioquímica de los aislados de P. dicentrarchi.
- Robledo Mahón, T. (2018). Estudio de los procesos biológicos y de la estructura de las comunidades microbianas en el proceso de compostaje de lodos de depuradora de aguas residuales urbanas, en sistemas de membranas semipermea.
- Coleto, I., Pineda, M., & Alamillo, J. M. (2019). Molecular and biochemical analysis of XDH from Phaseolus vulgaris suggest that uric acid protects the enzyme against the inhibitory effects of nitric oxide in nodules. Plant Physiology and Biochemistry.
- Álvarez Escribano, I. (2019). Caracterización de los sRNAs reguladores NsrR1 y NsiR3 de la cianobacteria Nostoc sp. PCC 7120.
- Moreno Cabezuelo, J. Á. (2019). Glucose uptake in marine cyanobacteria: regulation, expression of the transporter and effects on the proteome and metabolome.
- Alvarez, J. B., Castellano, L., Recio, R., & Cabrera, A. (2019). Wx Gene in Hordeum chilense: Chromosomal Location and Characterisation of the Allelic Variation in the Two Main Ecotypes of the Species. Agronomy, 9(5), 261.
- Benítez Casanova, L. (2017). Caracterización de las principales enzimas celulolíticas de myceliophthora thermophila implicadas en la degradación de biomasa lignocelulósica y mejora de la hidrólisis de hemicelulosa para la producción de bioetanol de segunda generación.
- Oliveras Rovira, À., Baró, A., Montesinos Barreda, L., Feliu Soley, L., & Planas i Grabuleda, M. (2018). Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens. PLoS ONE, 2018, vol. 13, núm. 7, p. e0201571.
- Folgueira, I., Lamas, J., De Felipe, A. P., Sueiro, R. A., & Leiro, J. M. (2019). Evidence for the role of extrusomes in evading attack by the host immune system in a scuticociliate parasite. BioRxiv, 607622.
- Olmos, G., Martínez‐Miguel, P., Alcalde‐Estevez, E., Medrano, D., Sosa, P., Rodríguez‐Mañas, L., … & López‐Ongil, S. (2017). Hyperphosphatemia induces senescence in human endothelial cells by increasing endothelin‐1 production. Aging cell, 16(6), 1300-1312.
- Laguna, J. I. R. (2017). Proliferative and Positional Instructions Underliying Planarian Regeneration and Tissue Renewal(Doctoral dissertation, Universitat de Barcelona).
- Lindo, L., McCormick, S. P., Cardoza, R. E., Kim, H. S., Brown, D. W., Alexander, N. J., … & Gutierrez, S. (2019). Role of Trichoderma arundinaceum tri10 in regulation of terpene biosynthetic genes and in control of metabolic flux. Fungal Genetics and Biology, 122, 31-46.
- Oliveras, À., Baró, A., Montesinos, L., Badosa, E., Montesinos, E., Feliu, L., & Planas, M. (2018). Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens. PloS one, 13(7), e0201571.
- Álvarez-Cao, M. E., Cerdán, M. E., González-Siso, M. I., & Becerra, M. (2019). Bioconversion of Beet Molasses to Alpha-Galactosidase and Ethanol. Frontiers in microbiology, 10.
- Álvarez-Escribano, I., Vioque, A., & Muro-Pastor, A. M. (2018). NsrR1, a nitrogen stress-repressed sRNA, contributes to the regulation of nblA in Nostoc sp. PCC 7120. Frontiers in microbiology, 9, 2267.
- TC, G. Supplemental Material. González-‐Aguilera C. et al.
- Burnat, M., Picossi, S., Valladares, A., Herrero, A., & Flores, E. (2019). Catabolic pathway of arginine in Anabaena involves a novel bifunctional enzyme that produces proline from arginine. Molecular Microbiology.
- Camó, C., Bonaterra, A., Badosa, E., Baró, A., Montesinos, L., Montesinos, E., … & Feliu, L. (2018). Antimicrobial peptide KSL-W and analogues: Promising agents to control plant diseases. Peptides.
- Ruiz Beltrán, C., Nadal i Matamala, A., Montesinos Seguí, E., & Pla i de Solà-Morales, M. (2018). Novel Rosaceae Plant Elicitor Peptides as sustainable tools to control Xanthomonas arboricola pv. pruni in Prunus spp. © Molecular Plant Pathology, 2018, vol. 19, núm. 2, p. 418-431.
- Ruiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., & Garrido-Ramos, M. A. (2017). Identification and Characterization of TALE Homeobox Genes in the Endangered Fern Vandenboschia speciosa. Genes, 8(10), 275.
- Ruiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., & Garrido-Ramos, M. A. (2017). Differential expression patterns of MIKCC-type MADS-box genes in the endangered fern Vandenboschia speciosa. Plant Gene, 12, 50-56.
- Aliaga‐Guerrero, M., Paullada‐Salmerón, J. A., Piquer, V., Mañanós, E. L., & Muñoz‐Cueto, J. A. (2018). Gonadotropin‐inhibitory hormone in the flatfish, Solea senegalensis: Molecular cloning, brain localization and physiological effects. Journal of Comparative Neurology, 526(2), 349-370.
- Muñoz-Jiménez, C., Ayuso, C., Dobrzynska, A., Torres-Mendéz, A., de la Cruz Ruiz, P., & Askjaer, P. (2017). An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. Genetics, 206(4), 1763-1778.
- Madroñal de Sancha, J. M. (2017). Estudios funcionales de las pirofosfatasas inorgánicas de saccharomyces cerevisiae y organismos fotosintéticos.
- Álvarez Cao, M. E. (2017). Optimización de la producción heteróloga de la enzima α-galactosidasa de” Saccharomyces cerevisiae” a partir de residuos agroindustriales.
- Atanasov, K. E., Liu, C., Erban, A., Kopka, J., Parker, J. E., & Alcázar, R. (2018). NLR mutations suppressing immune hybrid incompatibility and their effects on disease resistance. Plant Physiology, pp-00462.
- Reñé, A., Alacid, E., Figueroa, R. I., Rodríguez, F., & Garcés, E. (2017). Life-cycle, ultrastructure, and phylogeny of Parvilucifera corolla sp. nov.(Alveolata, Perkinsozoa), a parasitoid of dinoflagellates. European journal of protistology, 58, 9-25.
- Burnat, M., Li, B., Kim, S. H., Michael, A. J., & Flores, E. Homospermidine biosynthesis in the cyanobacterium Anabaena requires a deoxyhypusine synthase homologue and is essential for normal diazotrophic growth. Molecular Microbiology.
- Rengel, R., Smith, R. T., Haslam, R. P., Sayanova, O., Vila, M., & León, R. (2018). Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Research, 31, 183-193.
- Domínguez-Martín, M. A., López-Lozano, A., Clavería-Gimeno, R., Velázquez-Campoy, A., Seidel, G., Burkovski, A., … & García-Fernández, J. M. (2017). Differential NtcA responsiveness to 2-oxoglutarate underlies the diversity of C/N balance regulation in Prochlorococcus. Frontiers in Microbiology, 8, 2641.
- Calatrava, V., Hom, E. F., Llamas, Á., Fernández, E., & Galván, A. (2018). Ok, thanks! A new mutualism between Chlamydomonas and Methylobacteria facilitates growth on amino acids and peptides. FEMS Microbiology Letters.
- Velázquez-Palmero, D., Romero-Segura, C., García-Rodríguez, R., Hernández, M. L., Vaistij, F. E., Graham, I. A., … & Martínez-Rivas, J. M. (2017). An Oleuropein β-Glucosidase from Olive Fruit Is Involved in Determining the Phenolic Composition of Virgin Olive Oil. Frontiers in plant science, 8.
- Bornikoel, J., Carrión, A., Fan, Q., Flores, E., Forchhammer, K., Mariscal, V., … & Maldener, I. (2017). Role of Two Cell Wall Amidases in Septal Junction and Nanopore Formation in the Multicellular Cyanobacterium Anabaena sp. PCC 7120. Frontiers in cellular and infection microbiology, 7, 386.
- Prieto-Dapena, P., Almoguera, C., Personat, J. M., Merchan, F., & Jordano, J. (2017). Seed-specific transcription factor HSFA9 links late embryogenesis and early photomorphogenesis. Journal of experimental botany, 68(5), 1097-1108.
- Badosa, E., Montesinos, L., Camó, C., Ruz, L., Cabrefiga, J., Francés, J., … & Feliu, L. (2017). CONTROL OF FIRE BLIGHT INFECTIONS WITH SYNTHETIC PEPTIDES THAT ELICIT PLANT DEFENSE RESPONSES. Journal of Plant Pathology, 99, 65-73.
- De Felipe, A. P., Lamas, J., Sueiro, R. A., Folgueira, I., & Leiro, J. M. (2017). New data on flatfish scuticociliatosis reveal that Miamiensis avidus and Philasterides dicentrarchi are different species. Parasitology, 144(10), 1394-1411.
- Suarez‐Bregua, P., Chien, C. J., Megias, M., Du, S., & Rotllant, J. (2017). Promoter architecture and transcriptional regulation of musculoskeletal embryonic nuclear protein 1b (mustn1b) gene in zebrafish. Developmental Dynamics, 246(12), 992-1000.
- Ruiz, C., Nadal, A., Foix, L., Montesinos, L., Montesinos, E., & Pla, M. (2018). Diversity of plant defense elicitor peptides within the Rosaceae. BMC genetics, 19(1), 11.
- Martínez-Barriocanal, Á., Arcas-García, A., Magallon-Lorenz, M., Ejarque-Ortíz, A., Negro-Demontel, M. L., Comas-Casellas, E., … & Martín, M. (2017). Effect of Specific Mutations in Cd300 Complexes Formation; Potential Implication of Cd300f in Multiple Sclerosis. Scientific Reports, 7(1), 13544.
- Bornikoel, J., Carrión, A., Fan, Q., Flores, E., Forchhammer, K., Mariscal, V., … & Maldener, I. (2017). Role of Two Cell Wall Amidases in Septal Junction and Nanopore Formation in the Multicellular Cyanobacterium Anabaena sp. PCC 7120. Frontiers in cellular and infection microbiology, 7.
- Badosa, E., Montesinos, L., Camó, C., Ruz, L., Cabrefiga, J., Francés, J., … & Feliu, L. (2017). CONTROL OF FIRE BLIGHT INFECTIONS WITH SYNTHETIC PEPTIDES THAT ELICIT PLANT DEFENSE RESPONSES. Journal of Plant Pathology, 99.
- Suarez‐Bregua, P., Chien, C. J., Megias, M., Du, S. D., & Rotllant, J. (2017). Promoter architecture and transcriptional regulation of musculoskeletal embryonic nuclear protein 1b (mustn1b) gene in zebrafish. Developmental Dynamics.
- Munoz-Jimenez, C. M., Ayuso, C., Dobrzynska, A., Torres, A., de la Cruz-Ruiz, P., & Askjaer, P. (2017). An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. bioRxiv, 107029.
- Pilar Prieto-Dapena, Concepción Almoguera, José-María Personat, Francisco Merchan, Juan Jordano; Seed-specific transcription factor HSFA9 links late embryogenesis and early photomorphogenesis, Journal of Experimental Botany, Volume 68, Issue 5, 1 February 2017, Pages 1097–1108, https://doi.org/10.1093/jxb/erx020
- Ruiz-Roldán, C., Pareja-Jaime, Y., González-Reyes, J. A., & G. Roncero, M. I. (2015). The transcription factor Con7-1 is a master regulator of morphogenesis and virulence in Fusarium oxysporum. Molecular Plant-Microbe Interactions, 28(1), 55-68.
- Parejo-Farnés, C., Albaladejo, R. G., Arroyo, J., & Aparicio, A. (2013). A phylogenetic hypothesis for Helianthemum (Cistaceae) in the Iberian Peninsula/Una hipótesis filogenética para el género Helianthemum (Cistaceae) en la Península Ibérica. Botanica complutensis, 37, 83.
- Burnat, M., Schleiff, E., & Flores, E. (2014). Cell envelope components influencing filament length in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. Journal of bacteriology, 196(23), 4026-4035.
- Rodríguez, M. F. R., Sánchez-García, A., Salas, J. J., Garcés, R., & Martínez-Force, E. (2015). Characterization of soluble acyl-ACP desaturases from Camelina sativa, Macadamia tetraphylla and Dolichandra unguis-cati. Journal of plant physiology, 178, 35-42.
- Rodríguez-Rodríguez, M. F., Salas, J. J., Venegas-Calerón, M., Garcés, R., & Martínez-Force, E. (2016). Molecular cloning and characterization of the genes encoding a microsomal oleate Δ 12 desaturase (CsFAD2) and linoleate Δ 15 desaturase (CsFAD3) from Camelina sativa. Industrial Crops and Products, 89, 405-415.
- Parejo-Farnés, C. (2013). Una hipótesis filogenética para el género” Helianthemum”(” Cistaceae”) en la Península Ibérica. Botanica Complutensis, 37, 83-92.
- Olaya-Abril, A., Gómez-Gascón, L., Jiménez-Munguía, I., Obando, I., & Rodríguez-Ortega, M. J. (2012). Another turn of the screw in shaving Gram-positive bacteria: Optimization of proteomics surface protein identification in Streptococcus pneumoniae. Journal of proteomics, 75(12), 3733-3746.
- Chamizo-Ampudia, A., Galvan, A., Fernandez, E., & Llamas, A. (2011). The Chlamydomonas reinhardtii molybdenum cofactor enzyme crARC has a Zn-dependent activity and protein partners similar to those of its human homologue. Eukaryotic cell, 10(10), 1270-1282.
- Merino-Puerto, V., Herrero, A., & Flores, E. (2013). Cluster of genes that encode positive and negative elements influencing filament length in a heterocyst-forming cyanobacterium. Journal of bacteriology, 195(17), 3957-3966.
- Merino-Puerto, V., Herrero, A., & Flores, E. (2013). Cluster of genes that encode positive and negative elements influencing filament length in a heterocyst-forming cyanobacterium. Journal of bacteriology, 195(17), 3957-3966.
- Rodríguez-Rodríguez, M. F., Salas, J. J., Garcés, R., & Martínez-Force, E. (2014). Acyl-ACP thioesterases from Camelina sativa: Cloning, enzymatic characterization and implication in seed oil fatty acid composition. Phytochemistry, 107, 7-15.
- Olaya-Abril, A., Obando, I., & Rodríguez-Ortega, M. J. (2016). Data in support of proteomic analysis of pneumococcal pediatric clinical isolates to construct a protein array. Data in brief, 6, 917-922.
- Olaya-Abril, A., Jiménez-Munguía, I., Gómez-Gascón, L., Obando, I., & Rodríguez-Ortega, M. J. (2015). A pneumococcal protein array as a platform to discover serodiagnostic antigens against infection. Molecular & Cellular Proteomics, 14(10), 2591-2608.
- Olaya-Abril, A., Jiménez-Munguía, I., Gómez-Gascón, L., Obando, I., & Rodríguez-Ortega, M. J. (2013). Identification of potential new protein vaccine candidates through pan-surfomic analysis of pneumococcal clinical isolates from adults. PloS one, 8(7), e70365.
- Jiménez-Munguía, I., van Wamel, W. J., Olaya-Abril, A., García-Cabrera, E., Rodríguez-Ortega, M. J., & Obando, I. (2015). Proteomics-driven design of a multiplex bead-based platform to assess natural IgG antibodies to pneumococcal protein antigens in children. Journal of Proteomics, 126, 228-233.
- Burnat, M., & Flores, E. (2014). Inactivation of agmatinase expressed in vegetative cells alters arginine catabolism and prevents diazotrophic growth in the heterocyst‐forming cyanobacterium Anabaena. MicrobiologyOpen, 3(5), 777-792.
- Ejarque-Ortiz, A., Solà, C., Martínez-Barriocanal, Á., Schwartz Jr, S., Martín, M., Peluffo, H., & Sayós, J. (2015). The receptor CMRF35-like molecule-1 (CLM-1) enhances the production of LPS-induced pro-inflammatory mediators during microglial activation. PloS one, 10(4), e0123928.
- Munoz-Jimenez, C. M., Ayuso, C., Dobrzynska, A., Torres, A., de la Cruz-Ruiz, P., & Askjaer, P. (2017). An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. bioRxiv, 107029.
- Gómez-Marín, C., Tena, J. J., Acemel, R. D., López-Mayorga, M., Naranjo, S., de la Calle-Mustienes, E., … & Bovolenta, P. (2015). Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proceedings of the National Academy of Sciences, 112(24), 7542-7547.
- Rojas, M. (2014). Papel de los reguladores moleculares Fbp1 y Bmh2 en la virulencia de Fusarium oxysporum.
- VIDAL, A. G. (2016). Potencial de los tratamientos por termoterapia con agua caliente para el control de hongos de la madera de la vid y su efecto sobre la micoflora total, después de un año de cultivo.
- Corral Ramos, C. (2016). Metabolismo del glucógeno y procesos celulares implicados en dinámica nuclear y fusión de hifas en Fusarium oxysporum.
- Navarro Velasco, G. Y. (2013). Identificación de nuevos componentes de la ruta TOR de Fusarium oxysporum y determinación de su papel en la patogénesis.
- Ruiz-Roldán, C., Pareja-Jaime, Y., González-Reyes, J. A., & G. Roncero, M. I. (2015). The transcription factor Con7-1 is a master regulator of morphogenesis and virulence in Fusarium oxysporum. Molecular Plant-Microbe Interactions, 28(1), 55-68.
- Jiménez Munguía, I. (2015). Multi-omic approach applied to the selection of vaccine antigens and molecules for diagnosis and treatment agianst caused by Streptococcus pneumoniae and Staphylococcus aureus.
- Nürnberg, D. J. (2015). Intercellular communication in filamentous cyanobacteria
- Lambert Rodríguez, R. (2016). Actividad nucleasa en judía y su relación con la síntesis de ureidos durante la germinación y senescencia.
- Ocaña Calahorro, F. J. (2013). El óxido nítrico y la asmilación de nitrógeno en Chlamydomonas.
- Domínguez Martín, M. A. (2015). Diversity of regulatory mechanisms in the C/N metabolism of the marine cyanobacteria Prochlorococcus and synechococcus.
- Sein-Echaluce, V. C., Castejón, M. F. F., & Rodríguez, A. G. (2012). Estudios funcionales de la proteína FurB en Anabaena sp. PCC 7120.
- Bravo Ruiz, G. A. (2013). Sistemas hidrolíticos de componentes vegetales en el patógeno de tomate Fusarium oxysporum f. sp. lycopersici: lipasas y poligalaturonasas.
- Chamizo-Ampudia, A., Galvan, A., Fernandez, E., & Llamas, A. (2011). The Chlamydomonas reinhardtii molybdenum cofactor enzyme crARC has a Zn-dependent activity and protein partners similar to those of its human homologue. Eukaryotic cell, 10(10), 1270-1282.
Safety Statements
This product is developed, designed and sold exclusively for Research purposes and in vitro use only (RUO). The product was not tested for use in diagnostics or for drug development, nor is it suitable for administration to humans or animals. For more info, please check its Material Safety Data Sheet available in this website.
Customers Review
You may also like…
-

FastPANGEA™ High Fidelity DNA Polymerase
Price: 177.36 € P0032Add to basket -

T4 DNA Ligase (5U Weiss/µL)
Price: 118.52 € CL006Add to basket -

CVX5a™ Chemically Competent Cells
Price: 138.60 € – 345.00 € C0031Select options This product has multiple variants. The options may be chosen on the product page -

pSpark® I
Price: 175.00 € C0001Add to basket